| Description | Participants | Summaries | Products | Report |
|---|
Archived NIMBioS Working Group
Gene Tree/Species Tree Reconciliation
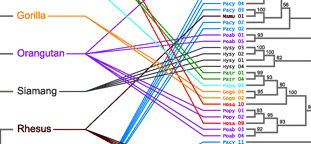
Topic: Inferring Patterns and Processes of Gene Diversification by Reconciling Gene Trees and Species Trees
Organizers:
Gordon Burleigh (Dept. of Biology, Univ. of Florida)
Oliver Eulenstein (Dept. of Computer Science, Iowa State Univ.)
David Liberles (Dept. of Molecular Biology, Univ. of Wyoming)
Meeting dates: December 16-18, 2010; August 10-12, 2011; November 26-29, 2012
Project summary: The wealth of genomic data across a broad phylogenetic spectrum provides an unprecedented opportunity to address one of the fundamental questions in evolutionary biology: what are the genomic mechanisms that generate species and phenotypic diversity? Gene duplication is critical for acquiring new gene functions and, consequently, adaptive innovations. Genomic surveys have revealed tremendous variation in gene content and copy number among species, and we are now challenged to place this variation in an evolutionary context. Gene tree and species tree reconciliation provides a direct method of inferring the patterns and processes of gene duplication and loss within the context of species evolution, as well as providing a rigorous method of gene tree rooting. Evolutionary processes, such as gene duplication and loss, lateral transfer, recombination, and incomplete lineage sorting, create incongruence between the phylogenies of genes and the species in which the genes evolve. Gene tree and species tree reconciliation problems seek to infer the evolutionary events that invoked the incongruence. While inferring the evolutionary history of gene families is a major concern for evolutionary genomicists, and algorithmic problems associated with gene tree and species tree reconciliation have long interested computer scientists, the work of these groups has rarely been fully integrated. The Gene Tree Reconciliation Working Group aims to define and characterize the statistical, algorithmic, and computational problems associated with gene tree and species tree reconciliation and to apply solutions to these problems to infer patterns and processes of gene duplication and loss from large-scale comparative genomic data sets.

Meeting Summaries
| Mtg # | Dates | Agenda | Summary | Photo | Evaluation |
|---|---|---|---|---|---|
| 1 | Dec 16-18, 2010 | Link | Link | Report | |
| 2 | Aug 10-12, 2011 | Link | Link | ||
| 3 | Nov 26-29, 2012 | Link |
Meeting 1 Summary. The primary goal of the first meeting was to summarize the progress in the field and identify critical areas for future research. We concluded that, although there have been many recent advances in gene trees-species trees (GT-ST) reconciliation from mathematicians, statisticians, computer scientists and biologists, these areas of research have rarely been fully integrated. The group defined several tractable projects that could benefit from the combined expertise of our group. These include statistical approaches for accessing models of gene evolution, methods for incorporating uncertainty and error in GT-ST analyses, and applications of GT-ST reconciliation for inferring the quality of genome annotations and plant genome evolution.
Meeting 2 Summary. The primary goal of the second meeting was to discuss progress from the first meeting and further develop our approaches for addressing gene tree reconciliation problems on biological data sets. Most of the first day was taken up with presentations and discussions of current research, including modeling gene evolution, identifying and accounting for error in gene tree topologies, phylogenetic approaches that account for gene coalescence, and modeling gene copy number. Biological applications for gene tree reconciliation approaches were also discussed. Based on these discussions and our previous work, we identified several major areas of focus: to include: 1) modeling complex processes of genomic evolution, 2) building more accurate gene trees and incorporating gene tree error in gene tree reconciliation methods, 3) phylogenetic inference based on processes of gene family evolution, and 4) identifying ancient whole genome duplications. An action plan for work to be done before the next meeting was developed, and tentative plans for the future directions of the working group were made.
Meeting 3 Summary. The primary goal of the final meeting was to cement new collaborations in several research trajectories and to discuss various levels of biological complexity in gene duplication and retention that are not currently considered. Strategies using both parsimony-based and model-based approaches were discussed, including computational aspects of identifying the optimal reconciliation and its relationship to the likelihood surface. Several members of the group will continue to work together on collaborations initiated at NIMBioS, including applications to international meeting hosts. Several collaborative proposals will also be sent to NSF and NIH.
 |
| Meeting 1 participants (Back row, L to R): P. Gorecki, L. Liu, M. Rasmussen, G. Burleigh, D. Liberles, O. Eulenstein, J. Leebens-Mack. (Front Row, L to R): C. Ane, A. Konrad, S. Huzurbazar, B. O'Meara, T. Jhwueng |
 |
| Meeting 2 participants (Back row, L to R): Oliver Eulenstein, Tony Jhwueng, Stefanie Hartmann, David Liberles, Gordon Burleigh; (Front, L to R): Jessica Wu, Brian O'Meara, Lars Arvestad, Snehalata Huzurbazar, Pawel Gorecki. Not pictured: Jim Leebens-Mack, Liang Liu |
NIMBioS Working Groups are chosen to focus on major scientific questions at the interface between biology and mathematics. NIMBioS is particularly interested in questions that integrate diverse fields, require synthesis at multiple scales, and/or make use of or require development of new mathematical/computational approaches. NIMBioS Working Groups are relatively small (up to 10 participants), focus on a well-defined topic, and have well-defined goals and metrics of success. Working Groups will meet up to 3 times over a two-year period, with each meeting lasting up to 2.5 days.
A goal of NIMBioS is to enhance the cadre of researchers capable of interdisciplinary efforts across mathematics and biology. As part of this goal, NIMBioS is committed to promoting diversity in all its activities. Diversity is considered in all its aspects, social and scientific, including gender, ethnicity, scientific field, career stage, geography and type of home institution. Questions regarding diversity issues should be directed to diversity@nimbios.org. You can read more about our Diversity Plan on our NIMBioS Policies web page. The NIMBioS building is fully handicapped accessible.
NIMBioS
1122 Volunteer Blvd., Suite 106
University of Tennessee
Knoxville,
TN 37996-3410
PH: (865) 974-9334
FAX: (865) 974-9461
Contact NIMBioS


