| Description | Participants | Summaries | Products |
|---|
NIMBioS Working Group
Ecological Network Dynamics
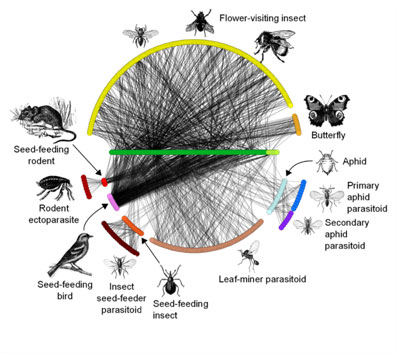
Topic: Spatiotemporal variation and dynamics in ecological networks
Meeting dates: Jun 10-12, 2015; Dec 7-11, 2015; Nov 14-17, 2016; Feb 13-15, 2019
Organizers:
David Hembry, Ecology and Evolutionary Biology, Univ. of Arizona
Dominique Gravel, Biology, Univ. de Sherbrooke, Quebec, Canada
Paulo Guimaraes Jr., Ecology, Univ. of Sao Paulo (USP), Brazil
James O'Donnell, School of Marine and Environmental Affairs, Univ. of Washington, Seattle
Objectives: The application of network analysis to ecological networks has made tremendous progress in the past two decades, greatly increasing our understanding of the assembly and dynamics of ecological communities. Likewise, the rapidly advancing field of spatial ecology has demonstrated that processes operating over spatial and temporal scales have strong effects on ecosystems and their constituent organisms. However, research in either field seldom incorporates information from the other. In part, this endeavor has been hindered by the limited availability of datasets spanning suitably large spatial or temporal scales. More problematic is the lack of a theoretical framework and the analytical tools needed to interpret the spatio-temporal dynamics of ecological networks. This working group brings together a diverse group of scientists whose expertise spans both fields, including field biologists along with theoreticians and computational biologists. The group includes empiricists working on a wide range of biological systems in order to ensure access to a broad array of data against which newly developed models are tested. Beyond facilitating discussion and collaboration among disparate groups, this group will develop working hypotheses for factors driving network dynamics based on empirical patterns; explore one or more case studies of variation across space or time in ecological networks; and develop a new model of network dynamics based on the theory of island biogeography, incorporating biogeography, coevolution, and community ecology, to be used in interpreting empirical patterns.

Meeting Summaries
| Mtg # | Dates | Agenda | Summary | Photo | Evaluation |
|---|---|---|---|---|---|
| 1 | Jun 9-12, 2015 | Link | Link | Report | |
| 2 | Dec 7-11, 2015 | Link | Link | ||
| 3 | Nov 14-17, 2016 | Link | Link | Report | |
| 4 | Feb 13-15, 2019 | TBA | Link | TBA |
Meeting 1 Summary. During our first meeting, we reviewed our general goal of better understanding the spatiotemporal dynamics of ecological networks, discussed gaps in the field, and identified areas where our group's strengths are best applied. Because ecological network analysis is generally removed from an evolutionary context, much of our discussion was centered on this theme. We established a plan for the longer-term goals of the group, and began work on four primary projects that emerged as tractable: 1) a macroevolutionary model of network evolution, 2) community assembly and evolution via discrete interaction rules, 3) scale dependence of ecological networks, and 4) a network model of extinction and invasion that incorporates behavior. While these four initial products primarily involve model development and simulation, we discussed existing empirical data sets, along with those that will be available at our next meeting.
Meeting 2 Summary. At our second meeting, the Working Group focused on two projects initiated at the first meeting: (1) a network model of extinction and (2) an extension of maximum information entropy methods to spatial network ecology. In project (1), we completed code development and debugging, preliminary analyses and their interpretation, and wrote a draft manuscript. In project (2), we wrote code for simulation of spatial networks and analysis of empirical data, through collaborative work among participants. Both projects advanced synthetic interactions among theoreticians and empiricists, in terms of incorporating biological realism into model parameterization and the identification of suitable datasets on which to test models. We also advanced projects initiated at the first meeting examining (3) scale dependence of ecological networks and (4) macroevolution and network evolution. We discussed via Skype the above and other projects from six participants unable to attend in person. Finally, we outlined several review/conceptual papers on different aspects of network analysis that we intend to produce as outcomes of this working group, and which we expect will significantly impact the field's efforts to synthesize mathematical and biological approaches to ecological network dynamics.
Meeting 3 Summary. At our third meeting, we focused on five projects which we initiated at earlier meetings: (1) a model of macroevolution and network evolution, (2) a simulation study investigating sampling bias from very large networks, (3) a network model of cascading extinction, (4) a model quantizing ecological interactions to investigate community assembly, and (5) a review of network analysis methods aimed at students and others new to the field. In project (1), we focused on code development and debugging, performed initial simulation runs, and wrote most of a first manuscript, as well as outlined goals for a second manuscript arising from the same model. In project (2), we continued code development and debugging, adding features which will allow the release of this code as a standalone Python software package that can be used by other scientists. We completed a draft for a first manuscript using this code to determine what kinds of biases arise when field researchers incompletely sample interactions from large networks of different structures. We also drafted a second manuscript presenting the new software package developed for project (2). In project (3), we identified and resolved problems with code developed during meeting 2, and completed code rewriting and debugging, as well as an outline for the future manuscript. In project (4), we completed code development, solicited feedback from all group participants, and wrote an outline for a first manuscript. In project (5), we received feedback and edits from all group members on a manuscript draft completed before the meeting, and expect to be able to submit the manuscript shortly. All projects advanced synthesis between computational and empirical approaches to the evolution of species interactions, especially through the identification of biologically interesting questions to be tested with each model.
 |
| Mtg. 1 participants (L to R): David Hembry, Justin Yeakel, Marie-Josee Fortin, Marcus Aloizio Martinez de Aguiar, Mathias Pires, Jimmy O'Donnell, Cecilia Diaz Castelazo, Erica Newman, Laura Burkle, Ulrich Mueller, Dominique Gravel, Timothee Poisot. |
 |
| Mtg. 2 participants (L to R): Cecilia Diaz Castelazo, Nels Johnson, David Hembry, Erica Newman, Marie-Josee Fortin, Laura Burkle, Jimmy O'Donnell. Not pictured: Ulrich Mueller, Timothee Poisot. |
 |
| Mtg. 3 participants (L to R): Justin Yeakel, Erica Newman, Jimmy O'Donnell, Marcus Aloizio Martinez de Aguiar, Mathias Pires, Timothee Poisot, Dominique Gravel, David Hembry, Laura Burkle, Marie-Josee Fortin. Not Pictured: Nels Johnson. |
 |
| Mtg. 4 participants. (L to R) Front: Mathilde Besson, Cecilia Diaz Castelazo, Mathias Pires, Laura Burkle, Jimmy O'Donnell; Rear: Justin Yeakel, Marcus Aloizio Martinez de Aguiar, David Hembry, Erica Newman, Ulrich Mueller. |
NIMBioS Working Groups are chosen to focus on major scientific questions at the interface between biology and mathematics. NIMBioS is particularly interested in questions that integrate diverse fields, require synthesis at multiple scales, and/or make use of or require development of new mathematical/computational approaches. NIMBioS Working Groups are relatively small (up to 10 participants), focus on a well-defined topic, and have well-defined goals and metrics of success. Working Groups will meet up to 3 times over a two-year period, with each meeting lasting up to 2.5 days.
A goal of NIMBioS is to enhance the cadre of researchers capable of interdisciplinary efforts across mathematics and biology. As part of this goal, NIMBioS is committed to promoting diversity in all its activities. Diversity is considered in all its aspects, social and scientific, including gender, ethnicity, scientific field, career stage, geography and type of home institution. Questions regarding diversity issues should be directed to diversity@nimbios.org. You can read more about our Diversity Plan on our NIMBioS Policies web page. The NIMBioS building is fully handicapped accessible.
NIMBioS
1122 Volunteer Blvd., Suite 106
University of Tennessee
Knoxville,
TN 37996-3410
PH: (865) 974-9334
FAX: (865) 974-9461
Contact NIMBioS


