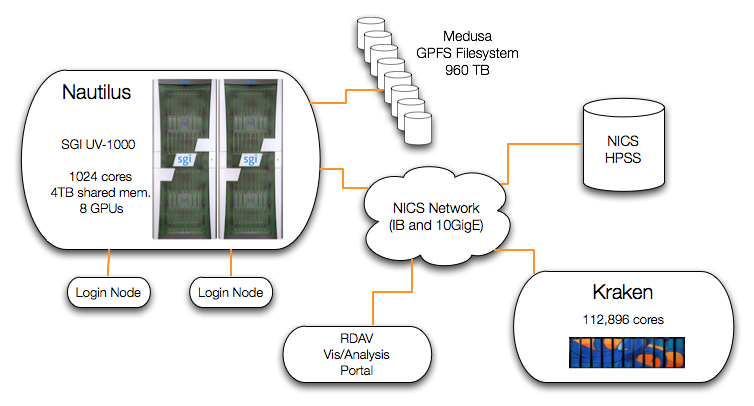
Nautilus Architecture
Nautilus is an SGI Altix UV1000 system and is the primary resource of the NICS Remote Data Analysis and Visualization Center (RDAV). It has 1024 cores (Intel Nehalem EX processors), 4 terabytes of global shared memory, and 8 GPUs in a single system image. Nautilus currently has a 960 TB GPFS file system, a CPU speed of 2.0 GHz, and a peak performance of 8.2 Teraflops. In addition, Nautilus is fully integrated with the NICS network which includes Kraken, Athena and the High Performance Storage System (HPSS).
Using Software via Modules
All software on Nautilus is managed through the module system. Here are descriptions of the various module commands:
module list- gives listing of currently loaded modulesmodule avail- shows all modules available on Nautilusmodule avail app- shows all modules available whose name matches app*module load app- sets up environment to use appmodule unload app- clears environment of references to appmodule swap app1 app2- unloads app1 and loads app2; usually used to swap versions or compilersmodule help app- gives specific usage info about appmodule whatis app- gives descriptive one-liner about app
The NICS webpage lists available software on Nautilus with descriptions and notes on usage:
Running Jobs via PBS
PBS options
Jobs are run on Nautilus through the Portable Batch System (PBS, also called Torque) which includes a job scheduler called Moab. Jobs can be run interactively or submitted to the scheduler as a batch script. When you submit a job (interactive or batch) you must supply PBS options which give the scheduler information about your run. Common options are:
-l ncpus=n- this one is mandatory; requests n cores for your job-l mem=m- specifies m memory for your job; defaults to 4GB for every core requested-l walltime=t- specifies time limit t for your job; defaults to 1 hour for interactive or batch jobs-A account- specifies account to charge for your job
See the NICS website for a complete list and more info on all PBS options:
Jobs are submitted using the qsub command as seen in the examples
below.
Interactive Job Example
To submit an interactive job, use the -I PBS option. For example:
qsub -I -l ncpus=8,walltime=3:00:00 -A UT-TENN0038
will request an interactive job using 8 cores for 3 hours for account
UT-TENN0038. When your job starts, you will be on a compute node on Nautilus
with the requested cores available. To end the interactive job, simply type
exit.
Batch Script Example
Here's an example PBS batch script to run an mpi program:
#PBS -A UT-TENN0033 #PBS -N mpi_job #PBS -j oe #PBS -q analysis #PBS -l ncpus=512,walltime=10:00:00 cd $PBS_O_WORKDIR mpiexec ./hwmpi
If this script is called script.pbs, you would submit it with:
qsub script.pbs
This script requests 512 cores and 10 hours of runtime. The additional PBS
options specify a name for the job (-N mpi_job),
the queue to run in (-q analysis) and the format for job
information output (-j oe
specifies one output file combining stdout and stderr).
$PBS_O_WORKDIR is an environment variable provided by PBS. It is
set to the path of the directory from which you executed qsub.
Let's say in this case that the hwmpi program is in ~/foo/bar. So from within
the ~/foo/bar directory, you issue qsub script.pbs. Now when your
job runs on a compute node, it will start in your home directory (~/). So
before it can run ./hwmpi successfully, it must cd to the directory ~/foo/bar--this is what
cd $PBS_O_WORKDIR accomplishes in the batch script.
Job Monitoring
Two helpful commands for monitoring your job's status are showq
and qstat. The qstat command generally gives more
detailed information about jobs in the system. You can use the -r
option with both of these commands to limit the information to only running
jobs.
Running Multiple Serial Jobs in Parallel
A Common Situation
Most of the time, code that is run through the batch system in an HPC
environment is parallel--either using MPI, OpenMP or pthreads. However, a
common situation is when a user needs to run multiple instances of some serial
code simultaneously to achieve parallelism.
On some systems, the mpiexec or mpirun command can be used to run non-MPI code as a
solution to this problem, however on Nautilus, this is not an option.
Here's a batch script that would achieve this on Nautilus:
#PBS -A UT-TENN0033
#PBS -N multi_job
#PBS -j oe
#PBS -q analysis
#PBS -l ncpus=512,walltime=10:00:00
cd $PBS_O_WORKDIR
./my_program &
./my_program &
./my_program &
. . .
./my_program &
wait
Essentially, this script will run my_program
512 times in the background
and then wait on all processes to finish. The operating system on Nautilus will
take care of actually distributing the processess over the 512 cores allocated.
While this approach is doable, there are some problems:
- What if you need to pass different command line arguments for each run of my_program? This can be very tedious to set up.
- What if you need to run 3000 instances and there are only 128 cores available?
- How will you manage the output from all of the different runs?
Enter Eden
We developed a tool called Eden as an answer to these problems. Eden makes it very easy to run as many serial jobs as you need on whatever resources are available.
Eden
Eden is entirely script-based (Bourne shell and some tcl) making it very portable and easy to use. To use Eden, all you need is a list of commands that you want to execute. This can be an explicit list that you generate yourself or you can give Eden a command template and a list of parameters from which Eden will generate the command list. Both ways are shown in the examples below.
Just as with any other software on Nautilus, load Eden through the module system:
module load eden
The basic usage of Eden is:
eden {commands|params} {PBS options}
The first argument tells Eden whether you're providing a list of commands or
a parameter file. This is followed by the PBS options for your job in the
same format as you would use for a qsub command when submitting an interactive
job.
Starting with Command List
Let's say you have the following commands that you want to run:
./my_program -t 0.2 ./my_program -t 0.4 ./my_program -t 0.6 ./my_program -t 0.8 ./my_program -t 1.0 ./my_program -t 1.2 ./my_program -t 1.4 ./my_program -t 1.6
First make a directory to use for your run.
Name your list of commands commands and place it in the directory.
Now you're ready to run Eden from your directory. This will be a very small example using only
4 cores to run the 8 commands:
module load eden eden commands -l ncpus=4,walltime=1:00:00 -A UT-TENN0033
Notice that the PBS options are in the same format as you would normally use with qsub. Eden will spit out some messages informing you of its actions and then submit your job to the queue. When your job is complete, you'll have a number of files in your directory:
commands_done- as commands are completed, their index number is written to this file; if a job is stopped prematurely, this file is used as a restart file for Eden; also, you can check this file as your job is running to monitor its progresseden.config- this file lists all of the information regarding your job; it is used to coordinate the different processess of Edeneden_job.pbs- this is the actual pbs script generated by Eden to submit your job- You'll also have the usual PBS output files from your job submission
In addition, you'll have a directory named with the timestamp of your Eden run.
Within that directory are all of the output files from the individual commands.
Each command will have a .out, a .err and a .time file.
Note that Eden treats each line of your commands file as a command
to run--but a single line could contain multiple commands, separated by
semicolons, if you wish.
Automatic Command Generation
Now let's say that your command list is going to be too much trouble to produce manually. You would rather just say, "Here's my program. Here are my parameters that I want to run through. Go." Eden makes this possible.
Here's an example params file that you would use (instead of the
commands file) to generate the command list from the above example:
./my_program -t $threshold threshold 0.2 0.4 0.6 0.8 1.0 1.2 1.4 1.6
Now you would call this file params and then run Eden with:
eden params -l ncpus=4
and Eden will automatically generate the command list to run your jobs.
Params File
The first line of the params file is your command template. This is
simply the command (or a series of commands separated by semicolons) that
you wish to run with placeholders for the
parameters that will vary. In the above example, $threshold is the
placeholder. Placeholders are signified by prepending a $ just as
you would for variables in a shell script.
Following the command template, you simply list the parameters (that correspond
to the placeholders) along with a list of values they can take.
This makes it very easy to accomplish runs in which you need to try all possible
permutations of a number of parameters. Here's another example params file:
./blackbox -t $threshold -p output$i.png $alpha $beta $gamma threshold 0.2 0.4 0.6 0.8 1.0 1.2 1.4 1.6 alpha NULL -a beta NULL -b gamma NULL -g
There are two additional things in this example. First of all, you can use the
built-in placeholder $i which is simply the index number of each
command. In this case, the output file signified by the -p
option will be appended with an incrementing index for each run. Also, if you
want a parameter to be either present or not present, use the NULL
keyword as one of the possible values for the parameter. Here are the first
few commands generated by the above params file:
./blackbox -t 0.2 -p output0.png ./blackbox -t 0.2 -p output1.png -g ./blackbox -t 0.2 -p output2.png -b ./blackbox -t 0.2 -p output3.png -b -g ./blackbox -t 0.2 -p output4.png -a ./blackbox -t 0.2 -p output5.png -a -g ./blackbox -t 0.2 -p output6.png -a -b ./blackbox -t 0.2 -p output7.png -a -b -g ./blackbox -t 0.4 -p output8.png
As you can see, Eden will generate every possible permutation of the values for threshold, alpha, beta and gamma.
Large-scale parameter studies with serial code can now be set up and managed quickly and easily with Eden on Nautilus.