Tutorial Calendar Accelerator Tutorials
Past NIMBioS Tutorials
Adaptive Management (Virtual)
 October 26-29, 2020.
Adaptive management seeks to determine sound management strategies in the face of uncertainty concerning the behavior of the system being managed. This tutorial introduced participants to methods for modeling adaptive management, with an emphasis on case studies drawn from environmental and natural resource management. The tutorial featured hands-on activities designed to help participants incorporate adaptive management approaches into their own research activities.
The tutorial was appropriate for faculty, post-docs, advanced graduate students, and industry professionals in resource economics, applied ecology, conservation biology, applied mathematics and related fields.
Some prior knowledge of dynamic optimization, population modeling and/or computer programming was recommended.
October 26-29, 2020.
Adaptive management seeks to determine sound management strategies in the face of uncertainty concerning the behavior of the system being managed. This tutorial introduced participants to methods for modeling adaptive management, with an emphasis on case studies drawn from environmental and natural resource management. The tutorial featured hands-on activities designed to help participants incorporate adaptive management approaches into their own research activities.
The tutorial was appropriate for faculty, post-docs, advanced graduate students, and industry professionals in resource economics, applied ecology, conservation biology, applied mathematics and related fields.
Some prior knowledge of dynamic optimization, population modeling and/or computer programming was recommended.
The Search for Selection
 June 3-7, 2019.
The NIMBioS Tutorial The Search for Selection
presented an integrated overview of the many approaches to searching for signatures of selection in organismal features, highlighting common themes and divergent assumptions.
It was applicable for population geneticists, genome biologists, evolutionary ecologists, paleontologists, functional morphologists, and just about any biologist who ponders on how to formally demonstrate that a feature (or features) of interest might have been shaped by selection.
This tutorial was a repeat of the NIMBioS tutorial offered June 18-22, 2018.
June 3-7, 2019.
The NIMBioS Tutorial The Search for Selection
presented an integrated overview of the many approaches to searching for signatures of selection in organismal features, highlighting common themes and divergent assumptions.
It was applicable for population geneticists, genome biologists, evolutionary ecologists, paleontologists, functional morphologists, and just about any biologist who ponders on how to formally demonstrate that a feature (or features) of interest might have been shaped by selection.
This tutorial was a repeat of the NIMBioS tutorial offered June 18-22, 2018.
Risk Assessment Calculator Training
 Mar 5-7, 2019.
The Institute for Environmental Modeling (TIEM) team has been publishing online "risk" (cancer risk and hazard index) calculators for over twenty years; this training provided the participant with operational knowledge of all TIEM calculators. Additionally, the instruction delved into the ability of the calculators to address site-specific exposures, unique toxicity assessments, and complex risk characterizations.
Mar 5-7, 2019.
The Institute for Environmental Modeling (TIEM) team has been publishing online "risk" (cancer risk and hazard index) calculators for over twenty years; this training provided the participant with operational knowledge of all TIEM calculators. Additionally, the instruction delved into the ability of the calculators to address site-specific exposures, unique toxicity assessments, and complex risk characterizations.
Network Modeling
 Feb 4-6, 2019.
This tutorial introduced students and interested faculty to the topic of complex networks. The field has grown tremendously over the last 20 years and network science has found numerous applications to fields such as biology, ecology, social sciences, physical sciences, computer science, technology, and urban planning. The tutorial reviewed in detail the main ideas and methods in network science with emphasis on both conceptual aspects and real-world applications. Hands-on activities helped the audience familiarize themselves with practical ways to potentially incorporate network analysis in their own research. No prior knowledge of networks or programming was required.
The tutorial was appropriate for interested faculty, post-docs, and graduate students of any discipline.
Feb 4-6, 2019.
This tutorial introduced students and interested faculty to the topic of complex networks. The field has grown tremendously over the last 20 years and network science has found numerous applications to fields such as biology, ecology, social sciences, physical sciences, computer science, technology, and urban planning. The tutorial reviewed in detail the main ideas and methods in network science with emphasis on both conceptual aspects and real-world applications. Hands-on activities helped the audience familiarize themselves with practical ways to potentially incorporate network analysis in their own research. No prior knowledge of networks or programming was required.
The tutorial was appropriate for interested faculty, post-docs, and graduate students of any discipline.
Applications of Spatial Data: Ecological Niche Modeling
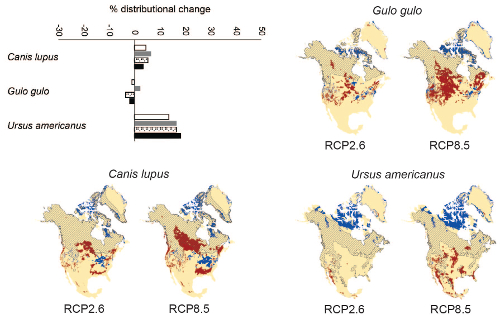 Dec 3-5, 2018.
The distribution of a species may be influenced by an array of factors. The set of conditions that allow a species to exist in a geographic area is a representation of the species ecological niche. However, defining these conditions is difficult, due to the complexity of natural systems. One approach to characterizing the ecological niche uses spatial data, GIS software, and modeling algorithms. The objectives of this tutorial were to teach participants the concepts of ecological niche modeling, introduce them to select analytical techniques (formatting GIS data; running maximum entropy – MaxEnt – models) and present how to interpret and apply spatial analyses. Participants were familiarized with several commonly-used and newly-available online spatial data resources. Instructors provided datasets to use in hands-on simulations, but participants could also bring their own data if desired. Applications of ecological niche modeling covered in this tutorial included biogeography, conservation biology, disease ecology, macroecology, and invasion biology.
Dec 3-5, 2018.
The distribution of a species may be influenced by an array of factors. The set of conditions that allow a species to exist in a geographic area is a representation of the species ecological niche. However, defining these conditions is difficult, due to the complexity of natural systems. One approach to characterizing the ecological niche uses spatial data, GIS software, and modeling algorithms. The objectives of this tutorial were to teach participants the concepts of ecological niche modeling, introduce them to select analytical techniques (formatting GIS data; running maximum entropy – MaxEnt – models) and present how to interpret and apply spatial analyses. Participants were familiarized with several commonly-used and newly-available online spatial data resources. Instructors provided datasets to use in hands-on simulations, but participants could also bring their own data if desired. Applications of ecological niche modeling covered in this tutorial included biogeography, conservation biology, disease ecology, macroecology, and invasion biology.
The Search for Selection
 June 18-22, 2018.
The NIMBioS Tutorial The Search for Selection
presented an integrated overview of the many approaches to searching for signatures of selection in organismal features, highlighting common themes and divergent assumptions.
It was applicable for population geneticists, genome biologists, evolutionary ecologists, paleontologists, functional morphologists, and just about any biologist who ponders on how to formally demonstrate that a feature (or features) of interest might have been shaped by selection.
June 18-22, 2018.
The NIMBioS Tutorial The Search for Selection
presented an integrated overview of the many approaches to searching for signatures of selection in organismal features, highlighting common themes and divergent assumptions.
It was applicable for population geneticists, genome biologists, evolutionary ecologists, paleontologists, functional morphologists, and just about any biologist who ponders on how to formally demonstrate that a feature (or features) of interest might have been shaped by selection.
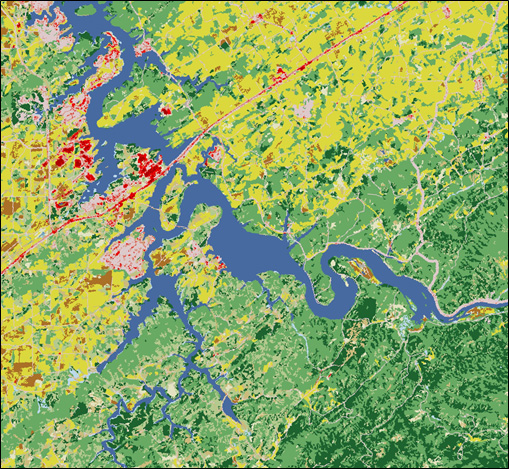
Applications of Spatial Data: Ecological Niche Modeling
May 16-18, 2018. NIMBioS hosted a tutorial
– Applications of Spatial Data: Ecological Niche Modeling.
The objectives of this tutorial were to teach participants the concepts of ecological niche modeling, introduce them to select analytical techniques (formatting data in GIS; running MaxEnt models), and present how to interpret and apply spatial analyses.
Phylogenetic Analysis Using RevBayes
Aug 7-11, 2017.
NIMBioS hosted an Accelerator Tutorial
– Phylogenetic Analysis Using RevBayes .
This tutorial featured RevBayes, an exciting new program for Bayesian inference of phylogeny that succeeds the popular program MrBayes. The course focused on phylogenetic trees and comparative-phylogenetic methods, including divergence-time estimation, morphological evolution, lineage diversification, and historical biogeography.
Uncertainty Quantification for Biological Models
June 26-28, 2017. NIMBioS hosted a tutorial –
Uncertainty Quantification for Biological Models.
This tutorial focused on uncertainty quantification in mathematical models in the life sciences and provided researchers with the basic concepts, theory, and algorithms necessary to quantify input and response uncertainties and to perform sensitivity analysis for simulation models.
Evolutionary Quantitative Genetics 2016
Aug 8-12, 2016. NIMBioS hosted a tutorial on Evolutionary Quantitative Genetics 2016, co-sponsored by the American Society of Naturalists. This tutorial was for evolutionary biologists interested in how quantitative genetics theory can be tested with data, both from single species and with multiple- species phylogenies. Participants - graduate students, postdocs, and junior faculty – learned how to use R to build and test evolutionary models.
Game Theoretical Modeling of Evolution in Structured Populations
April 25-27, 2016. NIMBioS hosted a tutorial on Game Theoretical Modeling of Evolution in Structured Populations, designed to introduce participants to the discrete graph theory methods and models of structured populations as well as classical continuous models based on differential equations.
Evolutionary Quantitative Genetics 2015
Aug 10-15, 2015. NIMBioS hosted a tutorial on Evolutionary Quantitative Genetics 2015, co-sponsored by the American Society of Naturalists.
NIMBioS/SAMSI/ESA Graduate Workshop:
April 15-17, 2015. NIMBioS hosted a workshop/tutorial on Current Issues in Statistical Ecology, designed to introduce and teach participants how to use computational resources for mathematical modeling and simulation of kinetic networks and networks in a spatial context.
Using R for HPC
NIMBioS hosted a tutorial Feb 27, 2015: An In-depth Introduction to Using R for HPC This tutorial was a joint training between the University of Tennessee, NIMBioS, XSEDE, and NICS.
Evolutionary Quantitative Genetics
NIMBioS hosted a tutorial Aug 4-9, 2014: Evolutionary Quantitative Genetics, co-sponsored by the National Evolutionary Synthesis Center (NESCent) and the American Society of Naturalists.
A BioQUEST Workshop at NIMBioS - Biology by Numbers: Bringing Math to the High School Biology Classroom
NIMBioS hosted a BioQUEST workshop/tutorial July 23-25, 2014 - Biology by Numbers: Bringing Math to the High School Biology Classroom.
Algebraic and Discrete Biological Models for Undergraduate Courses
NIMBioS sponsored a tutorial June 18-20, 2014: Algebraic and Discrete Biological Models for Undergraduate Courses.
Computing in the Cloud
NIMBioS sponsored a tutorial April 6-8, 2014: Computing in the Cloud: What Every Computational Life Scientist Should Know.
Parameter Estimation for Dynamic Biological Models
NIMBioS sponsored a tutorial May 19-21, 2014: Parameter Estimation for Dynamic Biological Models. This tutorial was designed for biologists interested in doing statistics with more complex non-linear models of their data and for mathematicians interested in learning how to apply their modeling skills to the unique demands of real dynamic biological data.
Mathematical Modeling for the Cell Biology Researcher and Educator
NIMBioS sponsored a tutorial April 8-10, 2013: Mathematical Modeling for the Cell Biology Researcher and Educator designed to introduce and teach participants how to use computational resources for mathematical modeling and simulation of kinetic networks and networks in a spatial context.
Using Bioinformatics Data and Tools to Engage Students in Problem Solving: A Curriculum Development Workshop
NIMBioS and Scalable Computing and Leading Edge Innovate Technologies (SCALE-IT) co-sponsored a workshop/tutorial Jan 7-10, 2013: Using Bioinformatics Data and Tools to Engage Students in Problem Solving: A Curriculum Development Workshop. The tutorial was co-presented by BioQUEST Curriculum Consortium and European Bioinformatics Institute (EBI).
Curriculum Development Faculty Workshop
NIMBioS and Scalable Computing and Leading Edge Innovate Technologies (SCALE-IT) co-sponsored a Curriculum Development Faculty Workshop/Tutorial, January 12-14, 2012, at NIMBioS, designed to support participants in the development of problem-based interdisciplinary computational biology courses in a curriculum that integrates cyber-learning strategies and data intensive activities into the classroom.
Multi-cell, Multi-scale Modeling
NIMBioS hosted a tutorial on Multi-cell, Multi-scale Modeling, May 18-21, 2011, at NIMBioS. This tutorial introduced and taught a novel, state-of-the-art approach to multi-cell multi-scale modeling using the Glazier-Graner-Hogeweg (GGH) and reaction kinetics network models and the CompuCell3D (CC3D) and Systems Biology Markup Language (SBML) simulation environments.
Migration from the Desktop: HPC application of R and other codes for biological research
NIMBioS hosted a tutorial on Migration from the Desktop: HPC application of R and other codes for biological research, May 9-11, 2011, at NIMBioS. The goal of this workshop was to introduce participants to the skills, strategies, and techniques necessary for them to make the jump from desktop computing to HPC environments.
Stochastic Modeling in Biology
NIMBioS hosted a tutorial on Stochastic Modeling in Biology, March 16-18, 2011, at NIMBioS. This tutorial was designed to introduce selected topics in stochastic modeling with an emphasis on biological applications.
Fast, Free Phylogenies: HPC for Phylogenetics
NIMBioS hosted a tutorial on High Performance Computing for Phylogenetics, Oct. 13-15, 2010, at NIMBioS. This tutorial was designed to teach participants how to use TeraGrid, the CIPRES Portal, the iPlant Discovery environment, university clusters, and other typically free HPC resources for phylogenetic analysis.
Graph Theory and Biological Networks
NIMBioS hosted a tutorial on Graph Theory and Biological Networks August 16-18, 2010, at NIMBioS. This tutorial was designed to teach participants how graph theory can inform their understanding of many common biological patterns that are graphs.
Computational Biology Curriculum Development
NIMBioS hosted a tutorial on Computational Biology Curriculum Development July 6-9, 2010, at NIMBioS. This tutorial focused on strategies for including computational biology and cyber-learning in the development of interdisciplinary modules for teaching undergraduate biology.
Optimization for Biologists
NIMBioS hosted a tutorial on Optimal Control and Optimization for Biologists Dec. 15-17, 2009, at NIMBioS. The tutorial introduced selected topics in optimal control and optimization with an emphasis on biological applications and included lectures and interactive computer lab sessions.
Training the Trainers
NIMBioS hosted a tutorial on High-Performance Computing for Computational Science Professionals Collaborating with Biologists March 16-18, 2009.
A goal of NIMBioS is to enhance the cadre of researchers capable of interdisciplinary efforts across mathematics and biology. As part of this goal, NIMBioS is committed to promoting diversity in all its activities. Diversity is considered in all its aspects, social and scientific, including gender, ethnicity, scientific field, career stage, geography and type of home institution. Questions regarding diversity issues should be directed to diversity@nimbios.org. You can read more about our Diversity Plan on our NIMBioS Policies web page. The NIMBioS building is fully handicapped accessible.
NIMBioS
1122 Volunteer Blvd., Suite 106
University of Tennessee
Knoxville,
TN 37996-3410
PH: (865) 974-9334
FAX: (865) 974-9461
Contact NIMBioS


