| Description | Central Theme | Participants | Agenda (pdf) | Products |
|---|
NIMBioS Tutorial: Multi-cell, Multi-scale Modeling
Additional Information
Central Theme: To be realistic and predictive, biological models need to cover a broad range of scales – from intracellular (inside the cell) to multi-cell (between cells) to whole organs and beyond. During the last decade, a considerable effort producing models operating at the intracellular level, e.g. of genetic regulatory networks, has resulted in a number of tools for modeling network dynamics inside single cells. The most successful of these is the open-source Systems Biology Workbench (SBW) developed by co-Organizer, Dr. Sauro, and its associated SBML toolkits (including SOSLib). However, researchers have realized that tools which allow simulating multi-cell, multi-scale biological processes in-silico may aid progress in many areas, e.g., development of tumor therapies, regenerative medicine, or understanding aging.
|
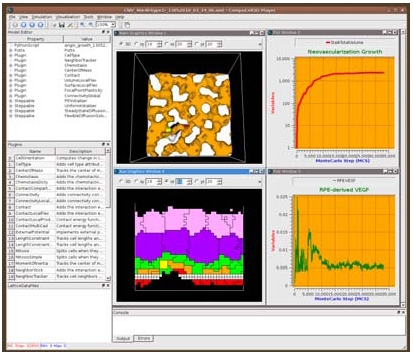
| Figure. 1: CC3D running a simulation of age-related macular degeneration. |
Very few problem solving environments support multi-cell modeling. CompuCell3D (CC3D), developed by Dr. Glazier, Dr. Swat and Mr. Heiland, is such a framework. It allows easy model definition, provides sophisticated model customization and allows linking of cell behaviors to underlying biochemical networks inside cells using SBW/SBML. Such models can span multiple scales – from intracellular to tissue level to organ. CC3D/SBW provides a versatile graphical user interface which makes visualization, post-processing and on-the-fly analysis of models easy and convenient. A snapshot of the CC3D application is shown in Figure 1.
A key feature of CC3D/SBW is that users define models using a multi-cell model-description language, CompuCell3D Markup Language (CC3DML), a comparable subcell language (Systems Biology Markup Language, SBML), and open-source Python scripting rather than a low-level language like C++ (hard coding) or a general purpose proprietary mathematical language like MATLAB or Mathematica. High-level model description reduces the knowledge of numerical analysis and programming required for model development and facilitates model publication, sharing, reuse and validation.
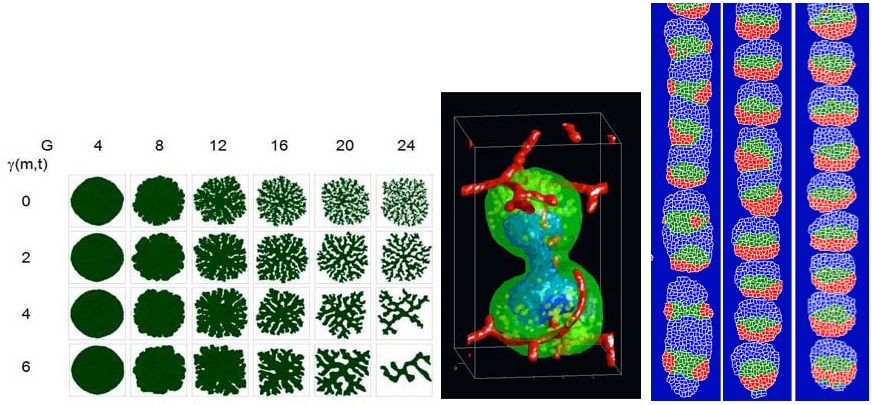
While the principles of coupled ODE and stochastic simulations of biochemical networks supported by SBW/SBML will probably be familiar to most researchers, the GGH multi-cell modeling approach may not be. The GGH is sufficiently simple that a simulation of 104 - 5×105 cells runs in a reasonable time on a single-processor laptop, but detailed enough to allow specification of cell shape-changes and movement, cell properties like volume, membrane area and type, and cell behaviors like adhesion, chemotaxis, polarization, growth, death and division. Its fixed lattice discretization makes it computationally simple and easy to interface with tissue-level continuum models of transport and diffusion and with subcellular models of regulation, metabolism and signaling. Unlike standard finite element (FE) methods, GGH allows the same simulation to run in 2D for speed or 3D for realism. It also supports FE-like, off-lattice overlays for situations where FE methods are helpful. Its advantages have led to its use by groups worldwide to simulate developmental phenomena from Dictyostelium discoideum aggregation and fruiting-body formation to vasculogenesis, omatidia patterning, and clot formation in blood vessels. A few sample models from CC3D are shown in Figure 2.
|
| Figure 2. CC3D simulations of: Tumor morphology as a function of nutrient consumption and cell adhesivity (left). Vascular tumor growing in a tissue with blood vessels (center). Somitogenesis model with successively more bio-realistic parameters (right). |
NIMBioS
1122 Volunteer Blvd., Suite 106
University of Tennessee
Knoxville,
TN 37996-3410
PH: (865) 974-9334
FAX: (865) 974-9461
Contact NIMBioS


